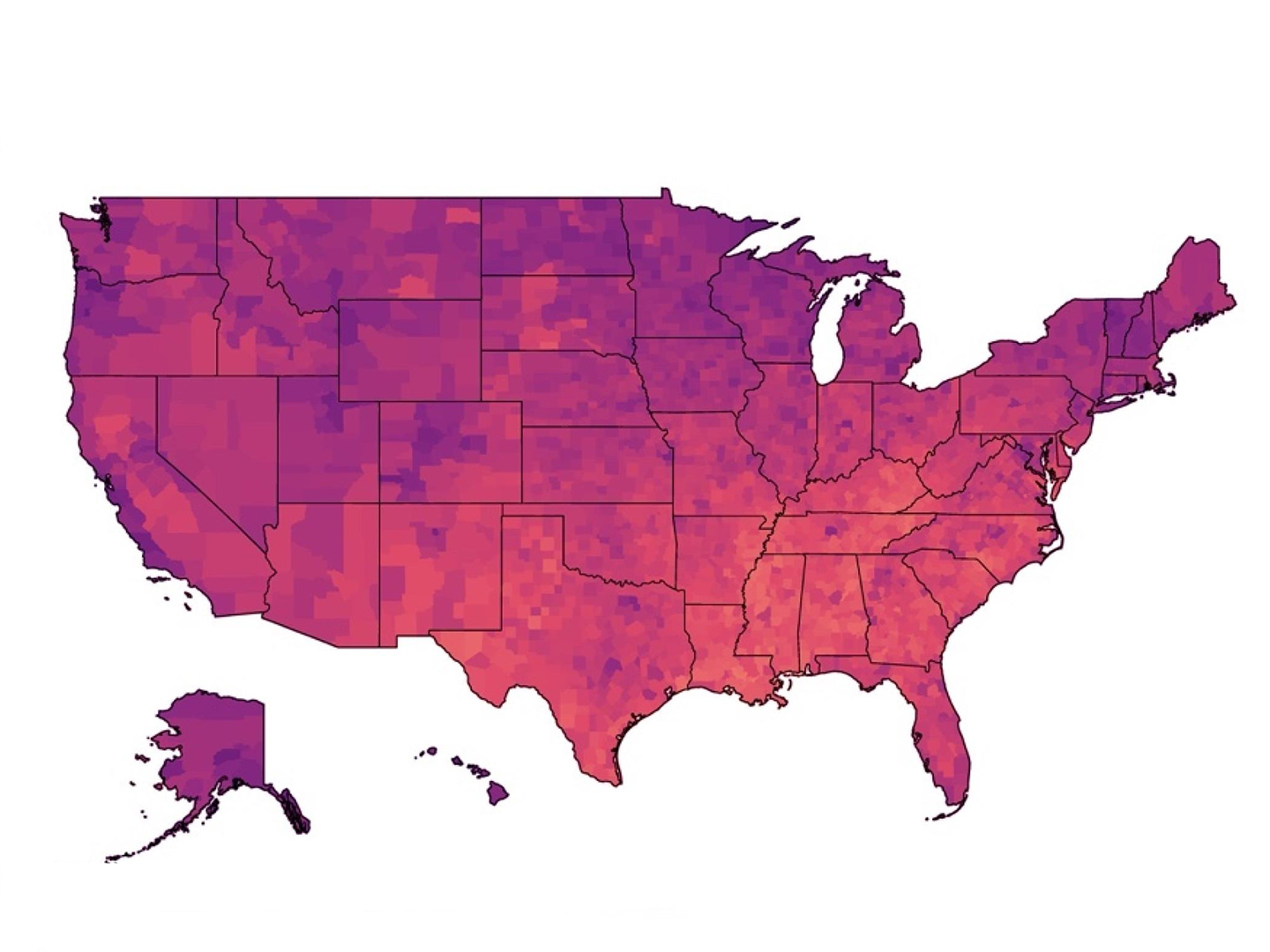
Climate Vulnerability Index Shows Where Action, Resources Are Needed To Address Climate Change Threats
Dr. Weihsueh Chiu, a professor at the Texas A&M School of Veterinary Medicine & Biomedical Sciences, partnered with Environmental Defense Fund to create a new tool that provides communities and policymakers with actionable data about long-term vulnerabilities tied to climate change. The Climate Vulnerability Index (CVI) is the most comprehensive screening tool of its type, showing […]
Genome Study Reveals 30 Years Of Darwin’s Finch Evolution
A landmark study on contemporary evolutionary change in natural populations released by an international team of researchers led by Texas A&M University professor Dr. Leif Andersson reveals that 45% of the variation in the highly heritable beak size of Darwin’s finches can be attributed to only six genomic loci (fixed positions on a chromosome). Among […]
Texas A&M Researchers Show Endangered Parrot Species Is Thriving In Urban Areas
A Texas A&M-led research team has discovered that a population of endangered red-crowned parrots is thriving in urban areas of South Texas. The parrots are a unique case, considering that many animal species are affected negatively by the expansion of human urban areas, which can lead to deforestation and pollution of natural habitats. These mostly […]
Texas A&M Veterinarians Developing Frailty Instrument To Improve Canine Geriatric Care
In human medicine, the ability to measure frailty is a vital aspect of geriatric care. Doctors may recommend one treatment over another based on an elderly person’s frailty score, and nursing homes may adjust care protocols as frailty increases. While the ability to measure frailty is not a new concept in human medicine, it has […]
Creating Partnerships Between Veterinarians, Ranchers To Enhance Profitability And Sustainability
Various Texas A&M faculty members partnered to create and deliver a workshop to selected areas within Texas to bring producers and veterinarians together. A research team from the Texas A&M School of Veterinary Medicine & Biomedical Sciences (VMBS) developed the first integrated, interdisciplinary food animal patient-client-veterinary relationship (PCVR) workshop available for use by continuing education […]
VMBS Doctoral Student Awarded For Research On Male Fertility
Pierre Ferrer, a Texas A&M School of Veterinary Medicine & Biomedical Sciences (VMBS) doctoral student, has been recognized with the American Society of Andrology’s 2023 Lonnie D. Russell Merit Award for his research on male fertility. The Andrology Society of America in a top international scientific society supporting both medical and basic research on the […]
Southwest Regional Superfund Summit Aims To Strengthen Data Science Collaborations, Training
Three Superfund Research Centers funded by the National Institute of Environmental Health Sciences (NIEHS) recently came together to share their accomplishments and challenges in fulfilling the data management and analysis mandate of the NIEHS Superfund Program. Scientists from Texas A&M University, the University of New Mexico, and the University of Arizona met in Albuquerque for […]
Superfund Research Center Trainees Share ‘One Health’ Research Initiatives
Five trainees from the Texas A&M Superfund Research Center presented research at a webinar hosted by the Superfund Research Program’s (SRP) Student, Postdoc, and Alumni Network (SPAN). The students participated in the Three-Minute Flash Talks portion of the webinar, which supplements graduate and advanced training by allowing trainees to present research to their peers while […]
Newest VERO Faculty Member Brings Environmental Microbiology To Texas Panhandle
Dr. Lee Pinnell is one of the few faculty members in Texas A&M’s Veterinary Education, Research, & Outreach (VERO) program who did not grow up planning to work in veterinary medicine. Rather than being a veterinarian like many of his VERO colleagues, Pinnell is an environmental microbiologist who has spent his education and career studying […]
Parasitology Students Bring Home Conference Presentation Awards
Three Texas A&M School of Veterinary Medicine & Biomedical Sciences (VMBS) students were recognized at the American Association of Veterinary Parasitologists’ (AAVP) annual conference for their outstanding research presentations. Hannah Danks, a fourth-year veterinary student, won Second Place Best Poster Presentation by a Student for her research distinguishing types of Dictyocaulus lungworms in white-tailed deer. […]